Structural Biology
Revealing Architecture, Function, and Interaction
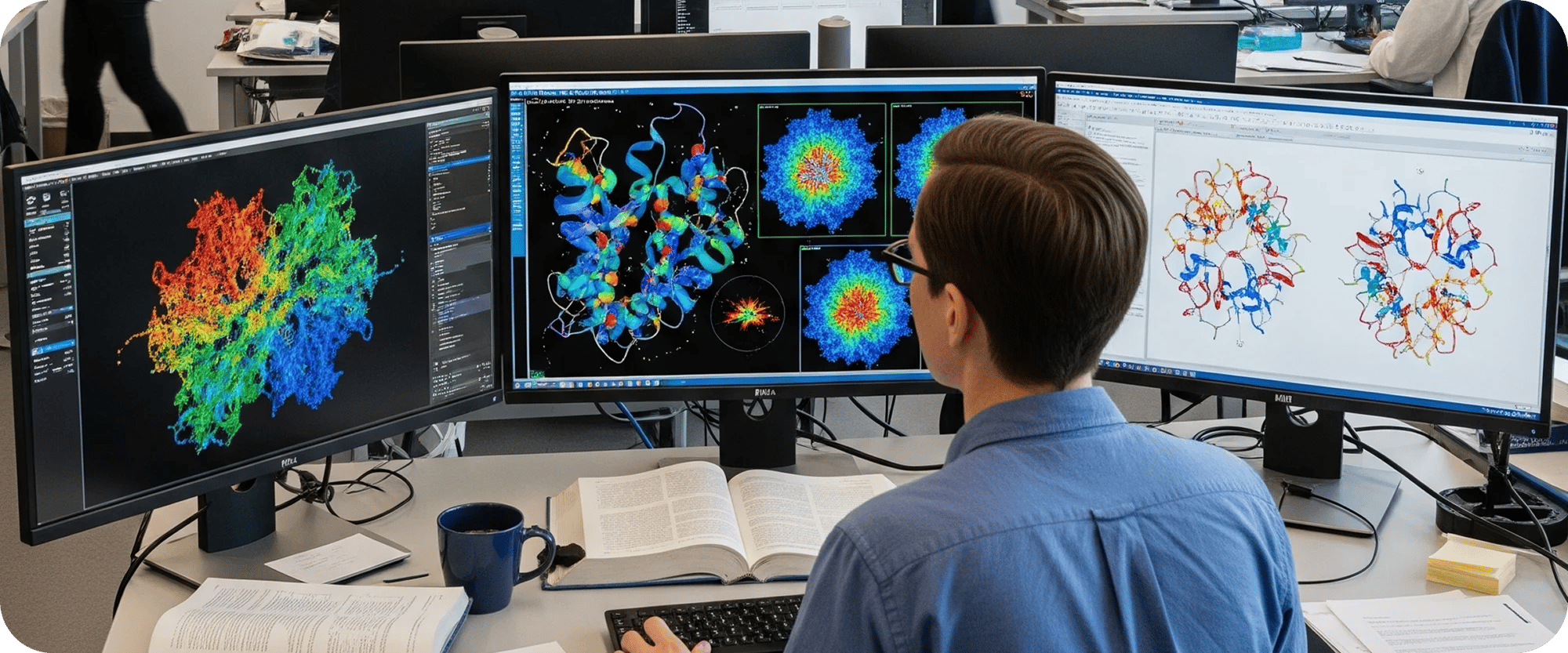
Structural biology is a fundamental discipline focused on determining the three-dimensional atomic and molecular structures of biological macromolecules, such as proteins, nucleic acids, and their complexes. These intricate structures are the very blueprints of life, dictating how molecules interact, catalyze reactions, and perform their myriad functions within a cell. Insights derived from structural biology are indispensable for rational drug design, understanding disease mechanisms, engineering novel biomolecules, and advancing our fundamental knowledge of biological processes. Complementing this, biophysical techniques provide crucial functional insights, quantifying molecular interactions, stability, and kinetics—essential context for understanding the biological relevance of a determined structure.
In recent years, breakthroughs in techniques like X-ray crystallography, Nuclear Magnetic Resonance (NMR) spectroscopy, and especially cryo-electron microscopy (cryo-EM) have revolutionized our ability to visualize biological structures at near-atomic resolution. Simultaneously, advancements in biophysical methods such as Surface Plasmon Resonance (SPR), Isothermal Titration Calorimetry (ITC), Nano Differential Scanning Fluorimetry (nanoDSF), and MicroScale Thermophoresis (MST) offer quantitative measures of binding kinetics, thermodynamics, and stability. Yet, harnessing these powerful experimental methods and translating raw data into meaningful structural models and functional insights presents significant computational and analytical challenges for organizations.
The Complexities of Structure, Interaction, and Data Integration
The journey from experimental data to a refined macromolecular structure and a comprehensive understanding of its biophysical behavior is computationally intensive and fraught with complexities. For cryo-EM, which has emerged as a dominant technique for large, flexible, and challenging complexes, the process involves acquiring vast amounts of noisy 2D images of individual particles. Processing these images to reconstruct a high-resolution 3D density map requires sophisticated algorithms for motion correction, contrast transfer function (CTF) estimation, particle picking, 2D classification, ab initio 3D reconstruction, and iterative 3D refinement. Each step demands specialized software, significant computational power, and meticulous parameter tuning.
A core challenge is dealing with protein dynamics and heterogeneity. Proteins are not static entities; they exist in multiple conformations and undergo dynamic changes crucial for their function. Capturing these different conformational states, understanding transient interactions, and characterizing structural changes during biological processes is extremely difficult. Traditional methods often yield static snapshots, making the interpretation of dynamic function challenging. Advanced structural biology requires computational approaches that can integrate data from multiple conformations or resolve heterogeneous structural populations.
Furthermore, integrating diverse experimental data sources—from high-resolution structural data to quantitative biophysical measurements—into a unified understanding is a complex task. The disparate data formats and the need for sophisticated algorithms to combine these heterogeneous inputs often hinder the creation of truly comprehensive insights. For instance, reconciling a determined X-ray or NMR structure with binding affinities (Kd), thermodynamic parameters (like ΔH from ITC), thermal denaturation temperatures (Tm from nanoDSF), or kinetic rates (Kon, Koff from SPR, or Kcat for enzyme kinetics) requires robust data pipelines and analytical frameworks.
Finally, while commercial software packages exist for various stages of structural and biophysical data processing, they often come with high licensing fees and may lack the flexibility or specialized features required for cutting-edge research on novel or difficult targets. The rapidly evolving nature of experimental techniques and computational modeling means that static, off-the-shelf solutions can quickly become outdated, forcing organizations into continuous, costly upgrades or limiting their research scope and the ability to fully leverage all available data.
Kupsilla's Expertise: Engineering Clarity in Macromolecular Architecture and Function
Kupsilla addresses these challenges by developing custom software solutions and providing expert services that streamline and enhance your structural biology and biophysical workflows. We empower your researchers to move efficiently from raw experimental data to high-resolution, functionally insightful molecular structures and quantitative interaction data, accelerating drug discovery, target validation, and fundamental biological understanding.
Our team possesses a profound understanding of the experimental methodologies of structural biology and biophysics, coupled with advanced computational and data engineering expertise. We are adept at developing solutions across the entire pipeline:
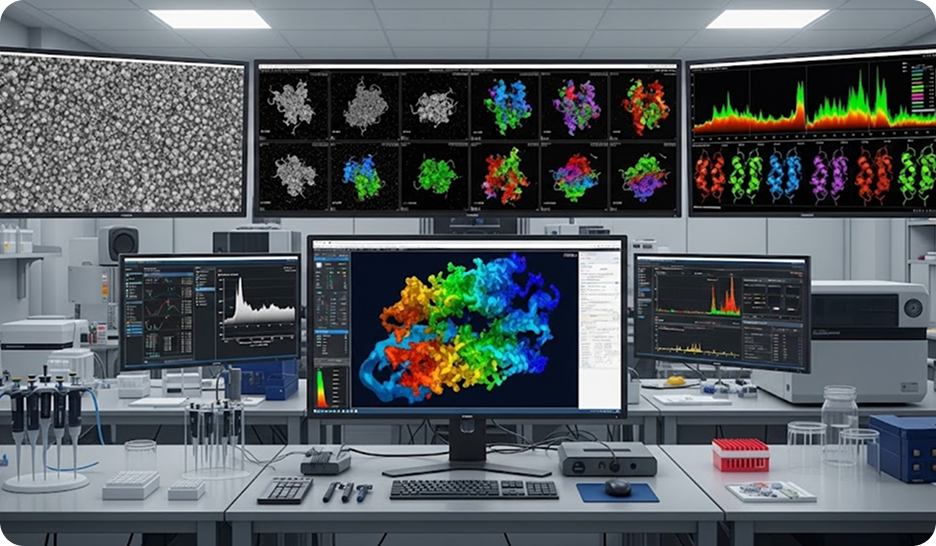
Custom Standardized Workflows for Data Processing
We design and optimize automated, standardized workflows for processing both biophysical (ITC, nanoDSF, MST, SPR) and structural biology (X-ray, NMR, cryo-EM) data. These robust pipelines ensure consistency, reproducibility, and high-throughput capabilities, from raw data acquisition to the derivation of key parameters like binding constants (Kd), melting temperatures (Tm), catalytic rates (Kcat), and diffusion coefficients (D).Cryo-EM Data Processing Pipelines
Designing and optimizing automated pipelines for high-throughput processing of cryo-EM data, including motion correction, CTF estimation, particle picking, 2D and 3D classification, ab initio reconstruction, and high-resolution refinement. We focus on enhancing speed, efficiency, and robustness for challenging datasets.Integrative Structural and Biophysical Modeling
Developing software to integrate data from multiple experimental techniques (e.g., cryo-EM maps, X-ray diffraction, NMR constraints, SAXS profiles) with quantitative biophysical data to build more complete and accurate models of macromolecular complexes, their interactions, and dynamic systems.
Protein Structure Prediction and Refinement
While AI tools like AlphaFold have revolutionized de novo prediction, we develop custom tools for model refinement, validation, and the prediction of multi-chain assemblies or specific protein-ligand interactions that require deeper computational insights beyond standard outputs.Conformational Dynamics Analysis
Building computational tools to analyze and visualize the dynamic behavior of macromolecules from simulation or experimental data, helping to understand flexibility, allostery, and reaction mechanisms.Structural and Biophysical Data Management and Visualization
Creating robust databases for managing structural models, biophysical curves and derived parameters, experimental metadata, and derived analyses, along with advanced visualization tools for interactive exploration and interpretation of complex structures, their dynamics, and their interaction profiles.
The Kupsilla Differentiator in Structural and Biophysical Informatics
What truly sets Kupsilla apart in the specialized field of structural and biophysical informatics is our commitment to delivering tailored, high-performance, and cost-effective solutions that provide true ownership:
- Mastery of Structural and Biophysical Data Processing: Our team has a strong grasp of the intricacies of structural biology data processing (particularly for cryo-EM, X-ray, and NMR) and the analysis of biophysical data (SPR, ITC, nanoDSF, MST). We understand the critical parameters, potential pitfalls, and optimization strategies for algorithms used in software like RELION, cryoSPARC, and EMready, as well as for extracting reliable results (e.g., Kd, Tm, Kcat, D) from biophysical experiments. We can develop custom scripts, interfaces, and automated workflows that extract maximum information from your experimental data, pushing the boundaries of resolution and interpretability.
- Expertise in Computer-Aided Drug Design (CADD) Tool Integration: We leverage structural and biophysical insights to empower your drug discovery efforts. Our expertise includes the integration and optimization of various CADD tools that can produce final results in different formats. This includes widely used open-source solutions like AutoDock Vina and AutoDock 4/GPU, as well as commercial packages such as Glide (HTVS, SP, and XP modes) and Flare by Cresset. We build custom workflows around these tools, enabling efficient virtual screening, lead optimization, and the identification of novel drug candidates based on precise structural information and validated binding characteristics.
- Expertise in Multi-Methodology Integration: We excel at integrating diverse computational and experimental methodologies to provide a holistic view of biological structures and their functional relevance. Whether it's combining cryo-EM with computational chemistry for ligand docking informed by biophysical binding data, or integrating predicted structures with experimental constraints from various sources, our solutions create seamless workflows that unlock insights not possible with single-technique approaches.
- Performance Optimization for HPC: Structural and biophysical computations are inherently demanding. We specialize in optimizing algorithms and workflows for high-performance computing (HPC) environments, including GPU acceleration and cloud-based solutions. This ensures that your complex reconstructions, simulations, and data analyses are executed with maximum speed and efficiency, significantly reducing turnaround times for critical structural and functional insights.
- Strategic Leverage of Open Source and Client Ownership: Kupsilla's core philosophy involves the strategic leveraging and enhancement of open-source software (e.g., Relion, ChimeraX, PyMOL, Rosetta, MDAnalysis, Biopython, statistical libraries for biophysics) that form the backbone of many structural and biophysical pipelines. We then customize, extend, and integrate these components with proprietary algorithms or commercial tools like Glide and Flare (where licensed) to create solutions precisely tailored to your research needs. This approach provides decisive advantages:
- Elimination of Escalating Licensing Costs for Custom Development: We free your organization from the burden of perpetual, per-user, or per-CPU licensing fees associated with custom software development and our services, focusing on value-driven, owned solutions.
- Complete Intellectual Property (IP) Ownership: Crucially, all the custom software, optimized pipelines, and unique algorithms we develop for you become your intellectual property. This eliminates vendor lock-in, provides unparalleled control over your structural and biophysical informatics infrastructure, and ensures that you can adapt, extend, and maintain your tools independently as your research evolves.
- Tailored for Unique Challenges: Our custom development approach ensures that the software is meticulously designed to address your specific structural and biophysical problems, whether it's processing challenging samples, integrating proprietary data, or implementing novel analytical methods not available in commercial packages.
By partnering with Kupsilla for your structural and biophysical informatics needs, you gain access to powerful, precise, and economically advantageous custom software solutions. We empower your scientists to accelerate the determination of crucial macromolecular structures, characterize their interactions, and leverage these insights for rational drug design, driving innovation in drug discovery and foundational research.